 The work by the researchers from Indiana University, University of Lausanne, Switzerland, Ecole Polytechnique Fédérale de Lausanne, Switzerland, and Harvard Medical School marks a major step in understanding the most complicated and mysterious organ in the human body. It not only provides a comprehensive map of brain connections (the brain “connectome”), but also describes a novel application of a non-invasive technique that can be used by other scientists to continue mapping the trillions of neural connections in the brain at even greater resolution, which is becoming a new field of science termed “connectomics.”
The work by the researchers from Indiana University, University of Lausanne, Switzerland, Ecole Polytechnique Fédérale de Lausanne, Switzerland, and Harvard Medical School marks a major step in understanding the most complicated and mysterious organ in the human body. It not only provides a comprehensive map of brain connections (the brain “connectome”), but also describes a novel application of a non-invasive technique that can be used by other scientists to continue mapping the trillions of neural connections in the brain at even greater resolution, which is becoming a new field of science termed “connectomics.”
“This is one of the first steps necessary for building large-scale computational models of the human brain to help us understand processes that are difficult to observe, such as disease states and recovery processes to injuries,” said Olaf Sporns, co-author of the study and neuroscientist at Indiana University.
The findings appear in the journal PLoS Biology today (June 30). Co-authors include Patric Hagmann and Reto Meuli, University Hospital Center and University of Lausanne; Leila Cammoun and Xavier Gigandet, Ecole Polytechnique Fédérale de Lausanne; Van J. Wedeen, Massachusetts General Hospital and Harvard Medical Center; and Christopher J. Honey, IU.
Until now, scientists have mostly used functional magnetic resonance imaging (fMRI) technology to measure brain activity — locating which parts of the brain become active during perception or cognition — but there has been little understanding of the role of the underlying anatomy in generating this activity. What is known of neural fiber connections and pathways has largely been learned from animal studies, and so far, no complete map of brain connections in the human brain exists.
In this new study, a team of neuroimaging researchers led by Hagmann used state-of-the-art diffusion MRI technology, which is a non-invasive scanning technique that estimates fiber connection trajectories based on gradient maps of the diffusion of water molecules through brain tissue. A highly sensitive variant of the method, called diffusion spectrum imaging (DSI), can depict the orientation of multiple fibers that cross a single location. The study applies this technique to the entire human cortex, resulting in maps of millions of neural fibers running throughout this highly furrowed part of the brain.
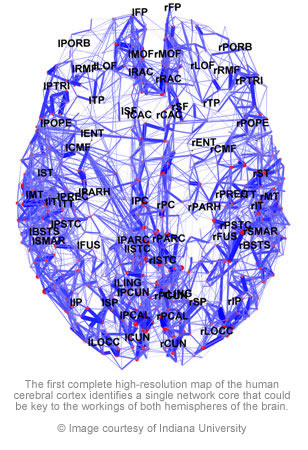
Sporns then carried out a computational analysis trying to identify regions of the brain that played a more central role in the connectivity, serving as hubs in the cortical network. Surprisingly, these analyses revealed a single highly and densely connected structural core in the brain of all participants.
“We found that the core, the most central part of the brain, is in the medial posterior portion of the cortex, and it straddles both hemispheres,” Sporns said. “This wasn’t known before. Researchers have been interested in this part of the brain for other reasons. For example, when you’re at rest, this area uses up a lot of metabolic energy, but until now it hasn’t been clear why.”
The researchers then asked whether the structural connections of the brain in fact shape its dynamic activity, Sporns said. The study examined the brains of five human participants who were imaged using both fMRI and DSI techniques to compare how closely the brain activity observed in the fMRI mapped to the underlying fiber networks.
“It turns out they’re quite closely related,” Sporns said. “We can measure a significant correlation between brain anatomy and brain dynamics. This means that if we know how the brain is connected we can predict what the brain will do.”
Sporns said he and Hagmann plan to look at more brains soon, to map brain connectivity as brains develop and age, and as they change in the course of disease and dysfunction.
The study can be viewed at http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0060159.
Source: Indiana University
Â
